What is Lactobacillus?
Lactobacillus is a genus of rod-shaped bacteria which, as the name suggests, produce lactic acid and thus belong to the Lactic Acid Bacteria (LAB) group. LAB are a mixed group of lactic acid-producing bacteria which require a fermentable carbohydrate for growth. Relationships between different LAB do not necessarily follow traditional classification schemes, however, they are all Gram-positive and non-sporing.
Lactobacillus species (spp.) can be divided into two types based on how they metabolise sugars: homofermentative and heterofermentative. Homofermentative Lactobacillus spp. ferment more than 90% of sugar to lactic acid and they do not produce gas. Heterofermentative species, on the other hand, ferment sugar into lactic acid but also substances such as acetic acid, and they produce CO2 gas. Although Lactic Acid Bacteria are associated with acid conditions regardless of whether they are Homo- or heterofermentative, both types need the pH to be above 3.2 for growth. Researchers developing probiotic products for skincare should note that heterofermentative Lactobacillus spp. will dominate a mixture of Lactobacillus spp. cultured at pH 5, i.e., around the pH of the skin. Further, remembering the outer stratum corneum layer of the epidermis can be quite dry, both types of Lactobacillus spp. prefer water activities (Aw) to be above 0.95 with fermentation stopping completely at Aw below 0.92 [2].
The taxonomy of LAB
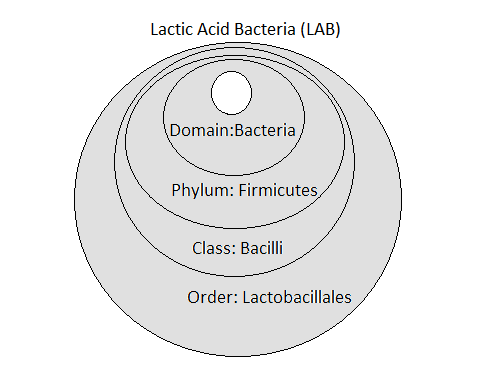
Figure 1 shows the taxonomy that most LAB have in common i.e., at the Domain, Phylum, Class and Order levels. At the next level (Genus) they can be cocci, coccobacilli or rods. Beyond Lactobacillus, LAB genera include: Aerococcus, Alloiococcus, Carnobacterium, Enterococcus, Lactococcus, Leuconostoc, Pediococcus, Streptococcus, Tetragenococcus, and Vagococcus.
Bifidobacteria are obligately anaerobic, Gram-positive bacteria, not classified with the LAB, but which occupy similar habitats and produce lactic acid as a sole end-product. The genus Bifidobacterium phylogenetically belongs to the phylum Actinobacteria, so they are only very distantly related to true LAB [1].
Lactobacillus bacteria are members of the order Lactobacillales, which are found in the class Bacilli. Bacilli is one of over 200 different classes belonging to the phylum Firmicutes.
Recent reclassification
Microbial classification can be confusing to the non-specialist. Classification is continuously undergoing subtle changes as more is learnt. However, taxonomy of Lactobacillaceae was really shaken up when DNA sequencing was applied in 2019. It is important therefore, when reading scientific papers on Lactobacillus spp. prior to this time, to first check where the bacteria species belongs in today’s taxonomy. Lactobacillales bacteria are fairly ubiquitous and abundant microbes; they are rod shaped, non-motile and aerotolerant anaerobes or microaerophilic. Until recently, the Lactobacillaceae family was believed to contain more than 250 Lactobacillus spp., however, in 2019, Lactobacillaceae was reclassified and reduced to nearer 25. The reclassification took place following whole genome sequence analysis, and was based on the microorganisms’ phylogenetic position, shared ecological and metabolic properties. The researchers involved point out that the generic term ‘lactobacilli’ can still remain useful, as a way to designate all organisms that were classified as Lactobacillaceae until 2019 [3].
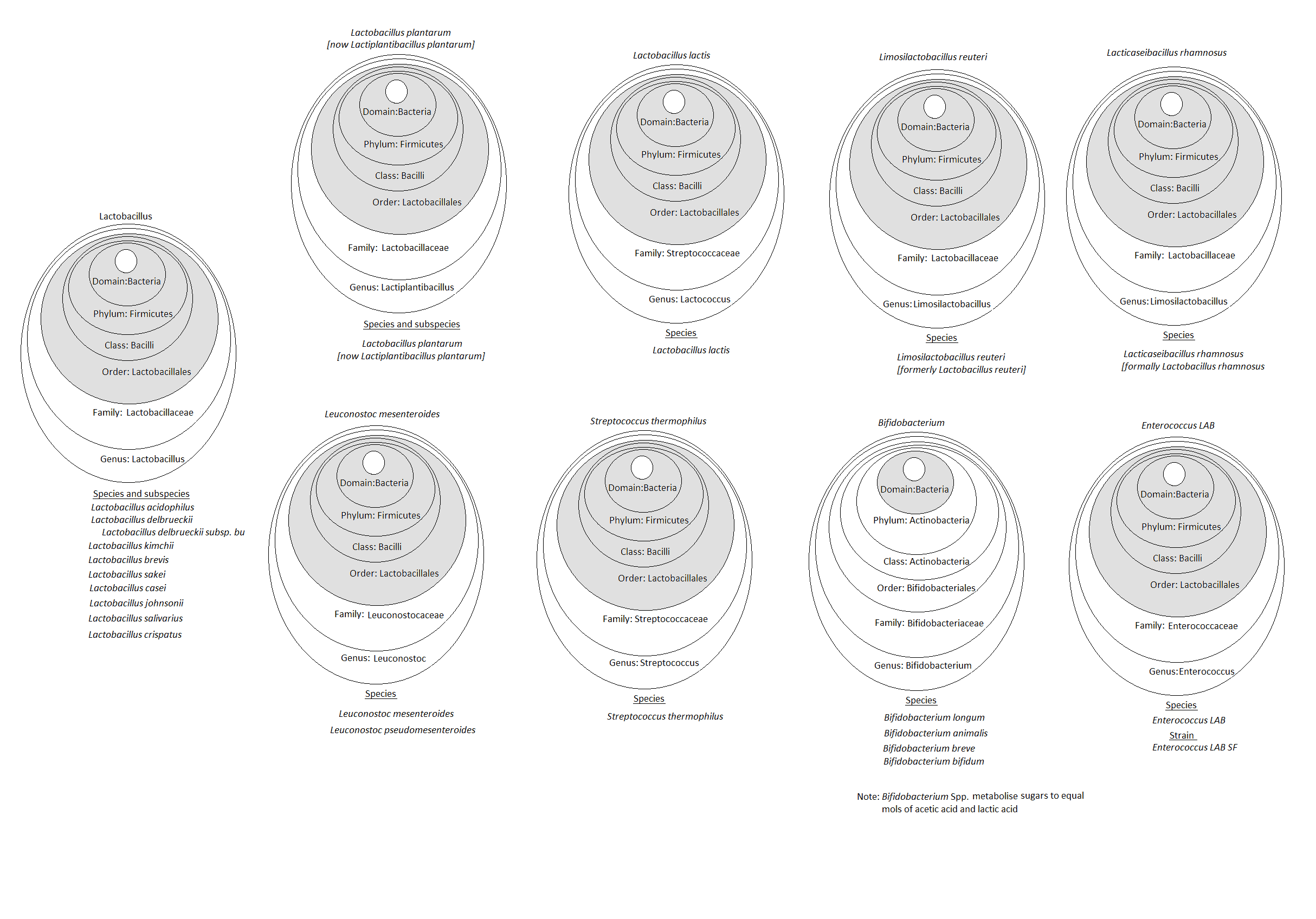
Figure 2 shows the taxonomy of many of the bacteria mentioned in this article and highlights the complexity of taxonomic classification. By shading the core taxonomy of Lactic Acid Bacteria (LAB), Domain, Phylum, Class and Order, the relationships between the different microbes, can be more easily seen.
Ubiquitous Lactobacillus
At the beginning of the 20th century, Martinus Beijerinck first identified Bacillus acidophilus (now known as Lactobacillus acidophilus), isolated from the human gut. Since then, Lactobacillus spp. have been found living in a wide variety of different habitats and hosts [4]. Most are found living with other Lactobacillus spp., for example, a polyphasic taxonomic approach revealed at least ten different strains of Lactobacillus coexisting in Chinese traditional pickle and yogurt [5]. Interestingly, nineteen strains of Lactobacillales were recently isolated from the International Space Station and its resupply vehicle [6].
LAB have relatively small genomes, which code more for acquiring carbon and nitrogen rich nutrients than for synthetic pathways. It is believed that through countless generations living in nutrient-rich environments, they have shed many unneeded genes for synthesis, while gaining genes for nutrient uptake through horizontal gene transfer [7].
Lactobacillus spp. along with other LAB bacteria, live at low levels on skin, whilst they overwhelmingly dominate healthy human vaginal microbiotas. These vaginal Lactobacilli benefit reproductive-age women by maintaining the pH at around 3.5–4.5 [8]. Besides creating a local acidic environment, Lactobacillus spp. influence microbiota by producing biofilms, providing nutrients, and also through releasing bacteriocins that help prevent potential pathogens from becoming established.
Lactobacillus as antimicrobials
Bacteriocins (and proteins with antimicrobial potential), produced by species such as Lactobacillus kimchii and strains of Latilactobacillus sakei are ”Generally Recognised as Safe” (GRAS) and so are of special interest as antimicrobials in food and cosmetics.
Latilactobacillus sakei produces sakacin P, which inhibits growth of several pathogenic and food-spoiling bacteria present in meat and fish products.
Over a decade ago, research was showing how the growth of Listeria monocytogenes in cold chicken cuts could be inhibited by the addition of sakacin P or sakacin P-producing Latilactobacillus sakei [9]. Possibly a better example is antimicrobial Nisin (Nisaplin® ) E234, Cas. 1414-45-5; EC No. 215-807-5, which is produced commercially from controlled Lactococcus Lactis fermentations of milk and other natural substrates. Nisin is listed on the EU cosmetics database (COSing) and has been used in foods since the 1960s. This and other bacteriocins are getting more attention now because of the current preservative crisis.
Bacteriocins are produced by one strain of bacteria working actively against a closely related strain, as part of their germ warfare approach to fighting for survival. It is only to be expected then to find that there areLeuconostoc mesenteroides strains, which produce bacteriocins to inhibit the growth of its microbial relative Latilactobacillus sakei [10].
Filtrates from cultures of Leuconostoc spp., grown in media with pulped radish root, can be found being used widely in cosmetics (Leuconostoc/radish root ferment filtrate), primarily for moisturising properties and secondarily for their accompanying broad-spectrum antimicrobial properties, which are believed to be due to bacteriocin(s). These antimicrobial properties are effective over the pH range 3-8 and at the conditions (temperatures and pHs) commonly used to manufacture cosmetics [11].
Lactobacillus and immunity
LAB cell surface components have been shown to prevent pathogens from infecting host cells and modulate the host’s immune system.
Several species of Lactobacillus e.g., Lactobacillus acidophilus, have an outer cell surface layer (S-layer) the functions of which are to protect the bacterium from aggressions, enable it to adhere to host or other surfaces, and to maintain its shape.
Lactobacillus acidophilus strains can have cell surface proteins (adhesins), such as S-layer protein A, which bind them to the same targets in the mucous, epithelium layers and extracellular matrix components in the human body, that pathogens adhere to.
Once bound, they prevent other microbes – including pathogens – from adhering, therefore exerting a protective effect. Some Lactobacillus acidophilus strains have murein hydrolases (lysins) in their surface S-layer. These hydrolytic enzymes digest the peptidoglycan layer (murein) of other bacterial cell walls, making them useful security guards against invading microbes [13].
Further, the interaction between LAB cell and host cell modulates the immune response by influencing the host’s dendritic cell and T cell functions. When a probiotic strain of Lactobacillus acidophilus adheres to dendritic cells in vitro there is a concentration-dependent production of anti-inflammatory (IL-10), and reduction in pro-inflammatory cytokine (il-12p70). IL-10 is important for maintaining gut homeostasis and its induction by Lactobacillus acidophilus may, in part, explain the anti-inflammatory effects of some probiotics [14]. Interestingly, Lactobacillus crispatus’ surface layer protein will work synergistically with Nisin by disrupting Staphylococcus saprophyticus cell walls enabling Nisin to form a lethal stable pore more easily [15].
Want to know more? Keep an eye out for part 2 to find out how these bacteria relate to probiotics, fermentation, and the microbiome. In the meantime, browse the Content hub for similar content – such as introducing E. coli parts 1 and 2.
References
[1] Pot B., Ludwig W., Kersters, K., Schleifer KH. (1994). ‘Taxonomy of Lactic Acid Bacteria’. In: De Vuyst L., Vandamme E.J. (eds) Bacteriocins of Lactic Acid Bacteria. Springer, Boston, MA. https://doi.org/10.1007/978-1-4615-2668-1_2
[2] Gerhard Feiner, G., (2006). ‘The microbiology of specific bacteria’, In Woodhead Publishing Series in Food Science, Technology and Nutrition, Meat Products Handbook, Ed. Feiner, G. Woodhead Publishing, Ch 39, Pages 595-615, ISBN 9781845690502,
https://doi.org/10.1533/9781845691721.3.595.
[3] Zheng, J., Wittouck, S., Salvetti, E., Franz, C.M.A.P., Harris, H.M.B., Mattarelli, P., O’Toole, P.W., Pot, B., Vandamme, P., Walter, J., Watanabe, K. (2020). ‘A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae’. International Journal of Systematic and Evolutionary Microbiology. 70 (4): 2782–2858. doi:10.1099/ijsem.0.004107. ISSN 1466-5026. PMID 32293557. https://doi.org/10.1099/ijsem.0.004107
[4] Bull, M., Plummer, S., Marchesi, J., and Mahenthiralingam, E., (2013) ‘The life history of Lactobacillus acidophilus as a probiotic: a tale of revisionary taxonomy, misidentification and commercial success’, FEMS Microbiology Letters, Volume 349, Issue 2, Pages 77–87, https://doi.org/10.1111/1574-6968.12293
[5] Long, G.Y., Wei Y.X., Tu, W., Gu C.T., (2020) ”Lactobacillus hegangensis sp. nov., Lactobacillus suibinensis sp. nov., Lactobacillus daqingensis sp. nov., Lactobacillus yichunensis sp. nov., Lactobacillus mulanensis sp. nov., Lactobacillus achengensis sp. nov., Lactobacillus mutagenesis sp. nov., Lactobacillus gannanensis sp. nov., Lactobacillus binensis sp. nov. and Lactobacillus angrenensis sp. nov., isolated from Chinese traditional pickle and yogurt” International Journal of Systematic and Evolutionary Microbiology. Volume 70, Issue 4 .
First Published: 26 February 2020 https://doi.org/10.1099/ijsem.0.004060
[6] Bharadwaj, A.R., Singh, K.N., Wood, J.M., Debieu, M., O’Hara, N.B., Karouia, F., Mason, C.E., Venkateswaran, K., (2020) ”Draft Genome Sequences of Lactobacillales Isolated from the International Space Station”. Microbiology Resource Announcements Sep 2020, 9 (39) e00942-20. DOI: 10.1128/MRA.00942-20
[7] Makarova, K., Slesarev, A., Wolf, Y., Sorokin, A., Mirkin, B., Koonin, E., Pavlov, Pavlova, A.N.,Karamychev, V., Polouchine, N., Shakhova,V., Grigoriev, I., Lou, Y., Rohksar, D., Lucas, S., Huang, K., Goodstein, D.M., Hawkins, T., Plengvidhya, V., Welker, D., Hughes, J.,Goh, Y., Benson, A., Baldwin, K.,Lee, J-H.,Díaz-Muñiz, I.,Dosti, B.,Smeianov, V. Wechter, W., Barabote, R.,Lorca, G., Altermann, E,Barrangou, R.,Ganesan, B.,Xie, Y.,Rawsthorne, H.,Tamir, D.,Parker, C.,Breidt, F.,Broadbent, J.,Hutkins, R.,O’Sullivan, D.,Steele, J.,Unlu, G.,Saier, M.,Klaenhammer, T.,Richardson, P.,Kozyavkin, S.,Weimer, B., and Mills, D.’Comparative genomics of the lactic acid bacteria’. Proc Natl Acad Sci U S A. 2006;103(42):15611-15616. doi:10.1073/pnas.0607117103
[8] Bradshaw C.S., Morton, A.N., Hocking J, Garland, S.M., Morris, M.B., Moss, L.M, Horvath, L.B., Kuzevska, I., Fairley, K.K., ”High recurrence rates of bacterial vaginosis over the course of 12 months after oral metronidazole therapy and factors associated with recurrence.” J. Infect. Dis. 2006;193:1478–1486.
[9] Katla, T., Møretrø, T., Sveen, I., Aasen. I.M., Axelsson, L., Rørvik, L.M., Naterstad, K. (2002). ‘Inhibition of Listeria monocytogenes in chicken cold cuts by addition of sakacin P and sakacin P‐producing Lactobacillus sakei‘. Vol. 93, Issue2. p 191-196.
[10] Kwang-Hee. L., and Jong-Hoon L., (2011) ‘Characterization of the Bacteriocin Produced by a Leuconostoc mesenteroides Strain Inhibiting the Growth of Lactobacillus sakei’. Microbiology and Biotechnology Letters . Volume 39 Issue 4. p390-396. https://www.koreascience.or.kr/article/JAKO201106736917180.page
[11] Schmitt E., and Norris, K., (2015). ‘Natural Alternatives for Cosmetic Preservation’. Happi (online) https://www.happi.com/issues/2015-02-01/view_features/natural-alternatives-for-cosmetic-preservation. Reprints available from More info: Active Concepts LLC, Lincolnton, NC, USA.
[12] Sfriso, R., Egert, M., Gempeler, M., Voegeli, R., and Campiche, R. (2019). ‘Revealing the secret life of skin ‐ with the microbiome you never walk alone.’ International Journal of Cosmetic Science. Volume42, Issue2. p116-126. https://doi.org/10.1111/ics.12594
[13] Acosta, M.P., Palomino, M.M., Allievi, M.C., Rivas, C.S., and Ruzal, S.M. (2008). ‘Murein Hydrolase Activity in the Surface Layer of Lactobacillus acidophilus ATCC 4356’ Applied and Environmental Microbiology Vol. 74 (24) p7824-7827; https://aem.asm.org/content/74/24/7824
[14] Konstantinov, S.R., Smidt, H., de Vos, W.M., Bruijns, S.C.M., Sing, S.K., Valence, F., Molle, D., Lortal, S., Altermann, E., Klaenhmmer, T.R., van Kooyk, Y. (2008). ‘S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions’. Proc Natl Acad Sci USA 105:1947–19479. https://doi.org/10.1073/pnas.0810305105
[15] Sun, Z., Li, P., Liu, F. et al. Synergistic antibacterial mechanism of the Lactobacillus crispatus surface layer protein and nisin on Staphylococcus saprophyticus . Sci Rep 7, 265 (2017). https://doi.org/10.1038/s41598-017-00303-8
